biotoolsSchema elements¶
A summary of elements in biotoolsSchema is below. For more detailed information, please see the technical documentation.
Important
When producing XML files compliant to the schema, it is essential to stick to the element order (including nested elements) as described below. See the sample XML files.
Element groups¶
biotoolsSchema includes scientific, technical and administrative software attributes, organised for convenience into 8 logical groupings (see below).
Note
As of biotoolsSchema 3.3.0, all of the element groups other than “Summary” and “Labels” are reflected as elements/objects in biotoolsSchema/biotoolsSchemaJ.
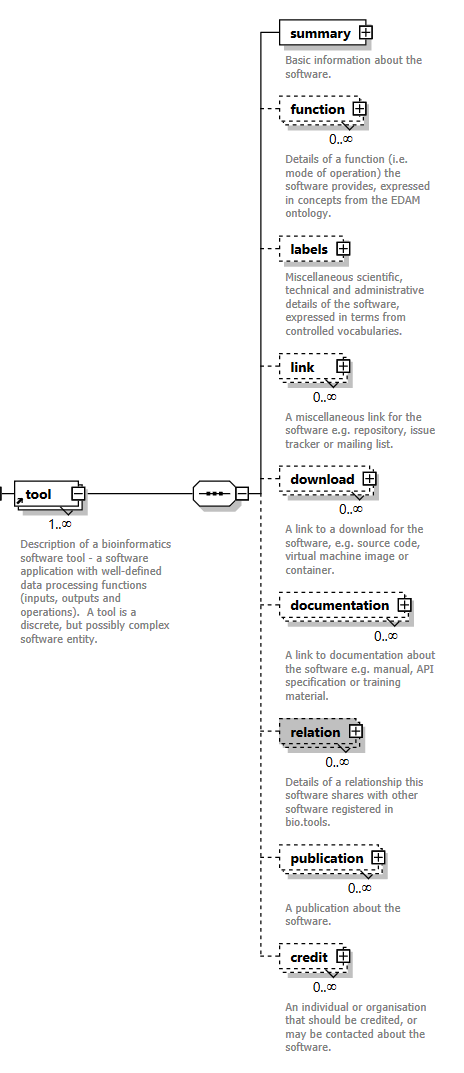
| Group | XSD element | Description |
|---|---|---|
| Summary | summary | Basic information about the software. |
| Function | function | Details of the function(s) (i.e. modes of operation) the software provides, expressed in terms from the EDAM ontology. |
| Labels | label | Miscellaneous scientific, technical and administrative details of the software, expressed in terms from controlled vocabularies. |
| Links | link | Miscellaneous links for the software e.g. repository, issue tracker or mailing list. |
| Downloads | download | Links to downloads for the software, e.g. source code, virtual machine image or container. |
| Documentation | documentation | Links to documentation about the software e.g. user manual, API documentation or training material. |
| Publications | publication | Publications about the software. |
| Relations | relation | Details of a relationship this software shares with other software registered in bio.tools. |
| Credits | credit | Individuals or organisations that should be credited, or may be contacted about the software. |
Summary group¶
Basic information about the software.
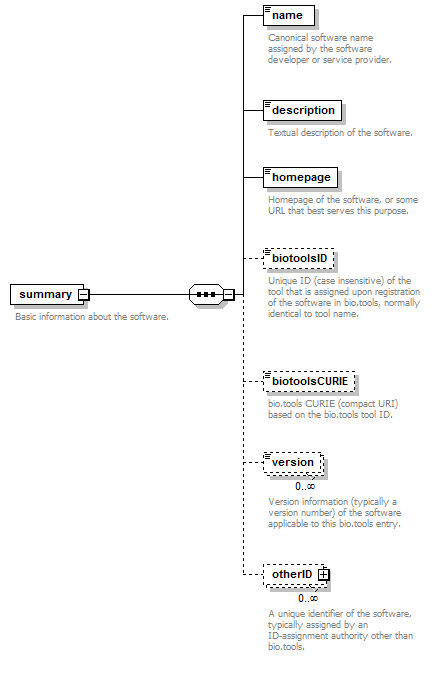
| Element | Description | Type | Cardinality |
|---|---|---|---|
| name | Canonical software name assigned by the software developer or service provider. | xs:token (restriction) | 1 only |
| description | Textual description of the software. | xs:token (restriction) | 1 only |
| homepage | Homepage of the software, or some URL that best serves this purpose. | URL | 1 only |
| biotoolsID | Unique ID (case insensitive) of the tool that is assigned upon registration of the software in bio.tools, normally identical to tool name. | URL (restriction) | 0 or 1 |
| biotoolsCURIE | bio.tools CURIE (compact URI) based on the bio.tools tool ID. | xs:token (restriction) | 0 or 1 |
| version | Version information (typically a version number) of the software applicable to this bio.tools entry. | xs:token (restriction) | 0 or more |
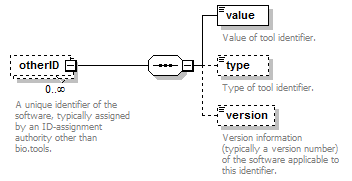
| otherID | A unique identifier of the software, typically assigned by an ID-assignment authority other than bio.tools. | (see below) | 0 or more |
| otherID->value | Value of tool identifier. | xs:token (restriction) | 1 only |
| otherID->type | Type of tool identifier. | enum (see docs) | 0 or 1 |
| otherID->version | Version information (typically a version number) of the software applicable to this identifier. | xs:token (restriction) | 0 or 1 |
Note
See the Curators Guide. As of biotoolsSchema 3.0.0, the Summary group does not have a corresponding element/object in biotoolsSchema/biotoolsSchemaJ (the schema was flattened).
Function group¶
Details of a function (i.e. mode of operation) the software provides, expressed in terms from the EDAM ontology.
Each software entity has one more functions, each corresponding to a mode of operation that the software provides. Each function performs one or more basic operations, and has zero or more primary input and/or output data, each of a specified type and supported format(s). For each operation, data or format element, an EDAM ontology concept URL and/or term are specified.
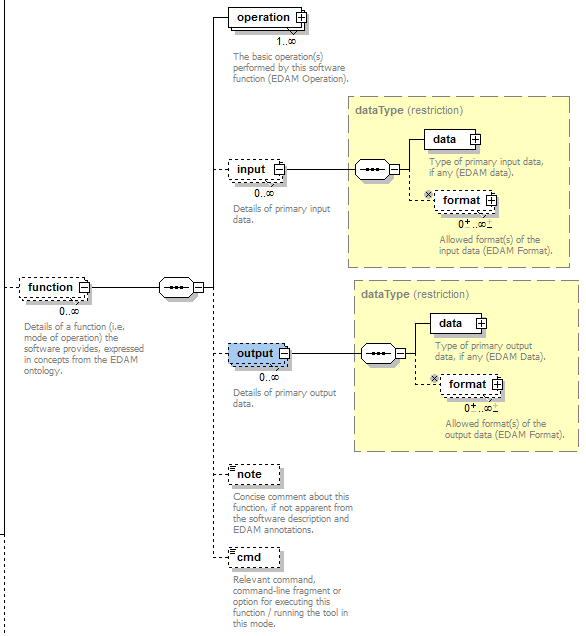
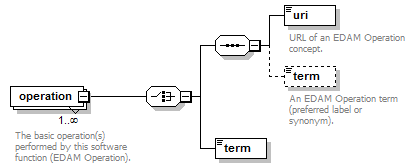
Data and Format are modelled in the same way as Operation (shown above).
| Element | Description | Type | Cardinality |
|---|---|---|---|
| operation | The basic operation(s) performed by this software function (EDAM Operation). | Ontology concept (restriction) | 1 or more |
| input | Details of primary input data. | ||
| input->data | Type of primary input data (EDAM data). | Ontology concept (restriction) | 1 only |
| input->format | Allowed format(s) of the input data (EDAM Format). | Ontology concept (restriction) | 0 or more |
| output | Details of primary output data. | ||
| output->data | Type of primary output data (EDAM Data). | Ontology concept (restriction) | 1 only |
| output->format | Allowed format(s) of the output data (EDAM Format). | Ontology concept (restriction) | 0 or more |
| operation | data | format->url | URL of an EDAM Operation | Data | Format concept. | URL (restriction) | 0 or 1 |
| operation | data | format->term | An EDAM Operation | Data | Format term (preferred label or synonym). | xs:token | 0 or 1 |
| note | Concise comment about this function, if not apparent from the software description and EDAM annotations. | xs:token (restriction) | 0 or 1 |
| cmd | Relevant command, command-line fragment or option for executing this function / running the tool in this mode. | xs:token (restriction) | 0 or 1 |
Important
The URL must be in the appropriate EDAM Operation | Data | Format namespace, e.g.
- http://edamontology.org/operation_0492
- http://edamontology.org/data_0863
- http://edamontology.org/format_1929
The term, e.g. “Multiple sequence alignment” must be either the preferred label of the concept or a synonym of this term, as defined in EDAM.
Note
See the Curators Guide.
Labels group¶
Miscellaneous scientific, technical and administrative details of the software, expressed in terms from controlled vocabularies.
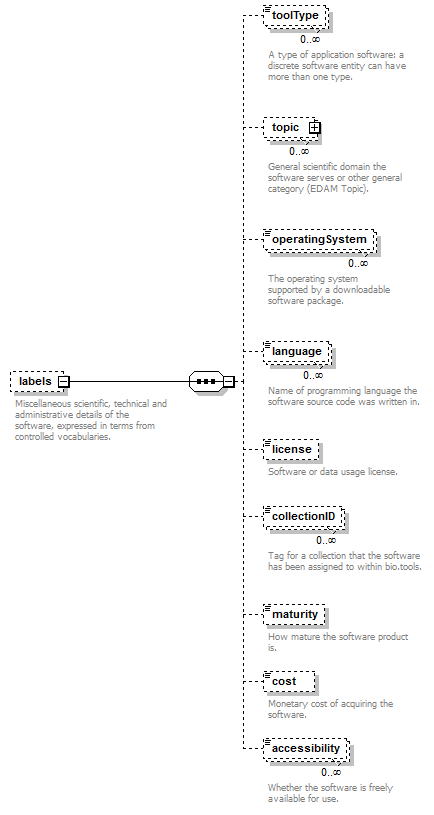
| Element | Description | Type | Cardinality |
|---|---|---|---|
| toolType | A type of application software: a discrete software entity can have more than one type. | enum (see docs) | 0 or more |
| topic | General scientific domain the software serves or other general category: one of EDAM Topic URL or term is specified. | Ontology concept (restriction) | 0 or more |
| topic->url | URL of an EDAM Topic concept. | URL (restriction) | 0 or 1 |
| topic->term | An EDAM Topic term (preferred label or synonym). | xs:token | 0 or 1 |
| operatingSystem | The operating system supported by a downloadable software package. | enum (see docs) | 0 or more |
| language | Name of programming language the software source code was written in. | enum (see docs) | 0 or more |
| license | Software or data usage license. | enum (see docs) | 0 or 1 |
| collectionID | Tag for a collection that the software has been assigned to within bio.tools. | xs:token (restriction) | 0 or more |
| maturity | How mature the software product is. | enum (see docs) | 0 or 1 |
| cost | Monetary cost of acquiring the software. | enum (see docs) | 0 or 1 |
| accessibility | Whether there are non-monetary restrictions on accessing an online service. | enum (see docs) | 0 or more |
| elixirPlatform | Name of the ELIXIR Platform that is credited. | enum (see docs) | 0 or more |
| elixirNode | Name of the ELIXIR Node that is credited. | enum (see docs) | 0 or more |
| elixirCommunity | Name of relevant ELIXIR (or associated) community. | enum (see docs) | 0 or more |
Note
See the Curators Guide. As of biotoolsSchema 3.0.0, the Labels group does not have a corresponding element/object in biotoolsSchema/biotoolsSchemaJ (the schema was flattened).
Link group¶
Miscellaneous links for the software e.g. repository, issue tracker or mailing list.
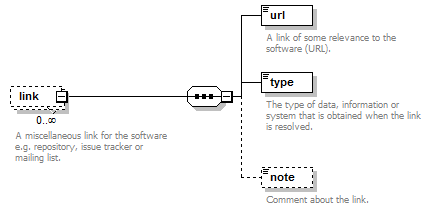
| Element | Description | Type | Cardinality |
|---|---|---|---|
| url | A link of some relevance to the software (URL). | URL | 1 only |
| type | The type of data, information or system that is obtained when the link is resolved. | enum (see docs) | 1 or more |
| note | Comment about the link. | xs:token (restriction) | 0 or 1 |
Note
See the Curators Guide.
Download group¶
Links to downloads for the software, e.g. source code, virtual machine image or container.
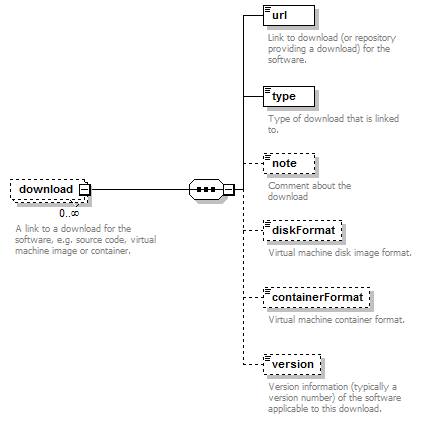
| Element | Description | Type | Cardinality |
|---|---|---|---|
| url | Link to download (or repo providing a download) for the software. | URL | 1 only |
| type | Type of download that is linked to. | enum (see docs) | 1 only |
| note | Comment about the download. | xs:token (restriction) | 0 or 1 |
| version | Version information (typically a version number) of the software applicable to this download. | xs:token (restriction) | 0 or 1 |
Note
See the Curators Guide.
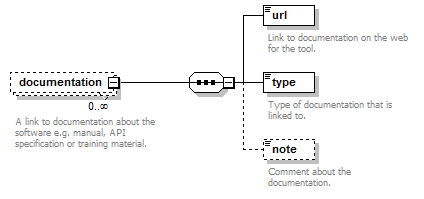
Documentation group¶
Links to documentation about the software e.g. manual, API specification or training material.
| Element | Description | Type | Cardinality |
|---|---|---|---|
| url | Link to documentation on the web for the tool. | URL | 1 only |
| type | Type of documentation that is linked to. | enum (see docs) | 1 or more |
| note | Comment about the documentation. | xs:token (restriction) | 0 or 1 |
Note
See the Curators Guide.
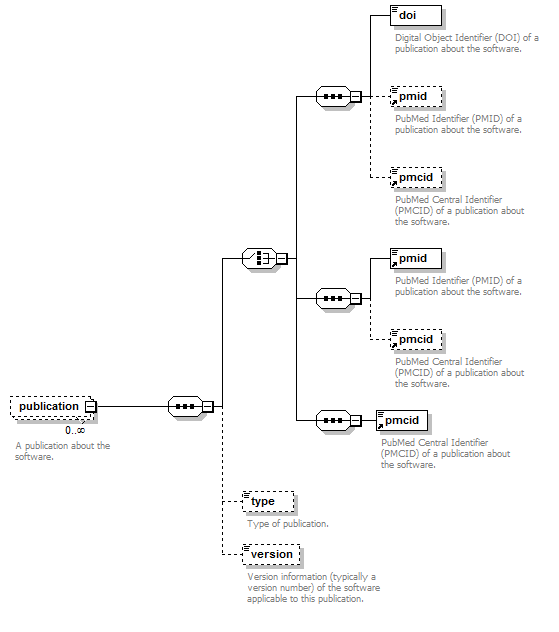
Publication group¶
Publications about the software
| Element | Description | Type | Cardinality |
|---|---|---|---|
| pmcid | PubMed Central Identifier of a publication about the software. | xs:token (restriction) | 0 or 1 |
| pmid | PubMed Identifier. | xs:token (restriction) | 0 or 1 |
| doi | Digital Object Identifier. | xs:token (restriction) | 0 or 1 |
| type | Type of publication. | enum (see docs) | 0 or more |
| note | Comment about the publication. | xs:token (restriction) | 0 or 1 |
| version | Version information (typically a version number) of the software applicable to this publication. | xs:token (restriction) | 0 or 1 |
Note
See the Curators Guide.
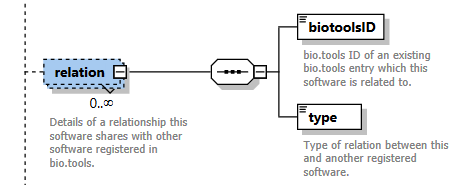
Relation group¶
Details of a relationship this software shares with other software registered in bio.tools.
| Element | Description | Type | Cardinality |
|---|---|---|---|
| biotoolsID | bio.tools ID of an existing bio.tools entry to which this software is related. | xs:token (restriction) | 1 only |
| type | Type of relation between this and another registered software. | enum (see docs) | 1 only |
Note
See the Curators Guide.
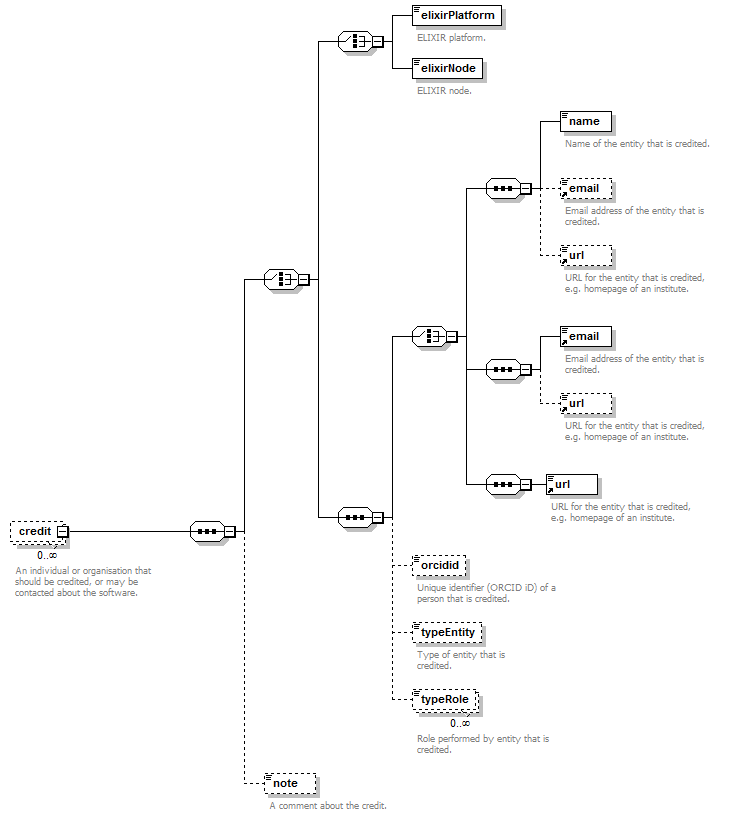
Credit group¶
Individuals or organisations that should be credited, or may be contacted about the software.
| Element | Description | Type | Cardinality |
|---|---|---|---|
| name | Name of the entity that is credited. | xs:token (restriction) | 0 or 1 |
| orcidid | Unique identifier (ORCID iD) of a person that is credited. | xs:token (restriction) | 0 or 1 |
| gridid | Unique identifier (GRID ID) of an organisation that is credited. | xs:token (restriction) | 0 or 1 |
| rorid | Unique identifier (ROR ID) of an organisation that is credited. | xs:token (restriction) | 0 or 1 |
| fundrefid | Unique identifier (FundRef ID or Funder ID) of a funding organisation that is credited. | xs:token (restriction) | 0 or 1 |
| Email address. | email address | 0 or 1 | |
| url | URL, e.g. homepage of an institute. | URL | 0 or 1 |
| tel | Telephone number. | xs:token (restriction) | 0 or 1 |
| typeEntity | Type of entity that is credited. | enum (see docs) | 0 or 1 |
| typeRole | Role performed by entity that is credited. | enum (see docs) | 0 or more |
| note | A comment about the credit. | xs:token (restriction) | 0 or 1 |
Note
See the Curators Guide.